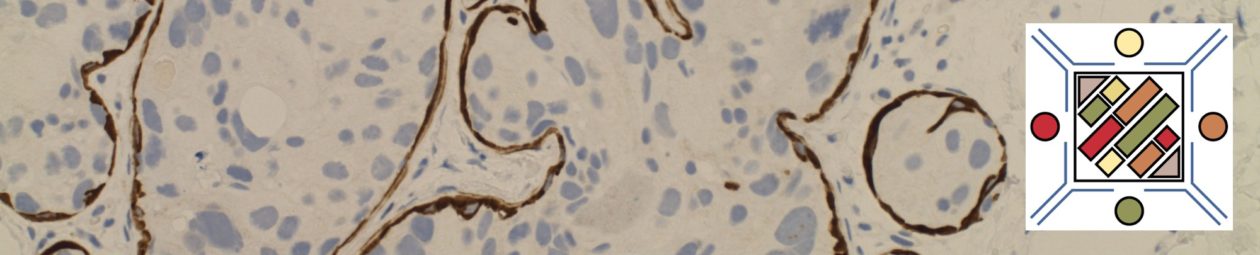
MUM-1/IRF-4 (multiple myeloma oncogene 1/Interferon Regulatory Factor-4) is a nuclear transcription factor, which is expressed in late stage germinal center cells (as bcl-6 expression begins to be down-regulated) and post-germinal center lymphocytes/plasma cells. Activated T-cells may also express MUM-1.
Tag Archives: MUM-1
Hans’ Algorithm
Diffuse Large B-Cell Lymphoma represents a heterogeneous group of non-Hodgkin B-cell lymphoma cases that share a common architectural pattern and large cell size. Subcategorization have been attempted with varying success based on morphology, immunophenotype, and molecular characteristics. Gene expression profiling (GEP) has demonstrated two important groups for both prognosis and treatment. Alizadeh, et al showed significant survival differences in cases of DLBCL with either a germinal center B-cell-like pattern or an activated B-cell-like pattern.
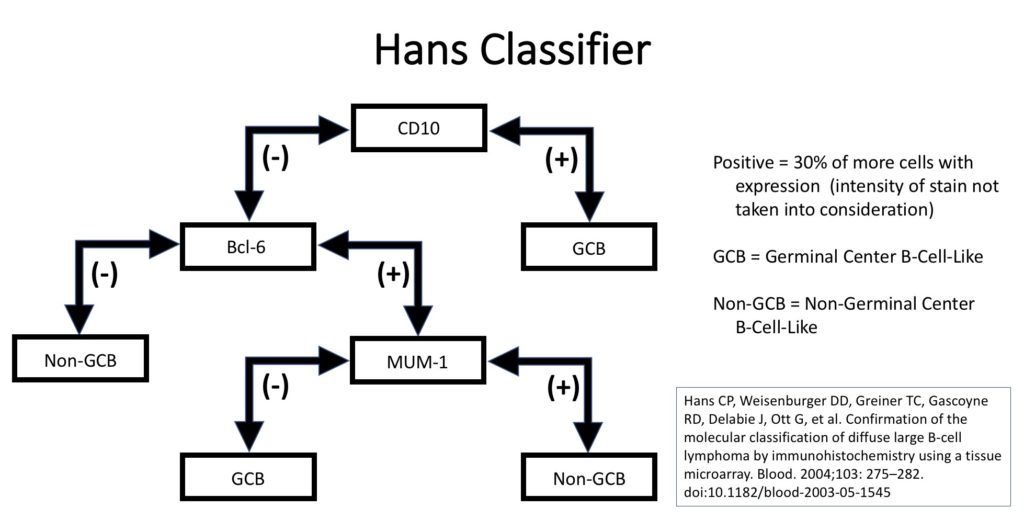
Unfortunately, GEP is not available in routine clinical practice, and multiple surrogate immunohistochemistry (IHC) based algorithms have been developed as a surrogate to GEP. The Hans’ algorithm (classifier) has been one of the most popular methods because it uses only three IHC markers (CD10, Bcl-6, & MUM-1) that are commonly available in most pathology laboratories. The following figure highlights the algorithm for the Hans’ classifier as described in the original paper. The Hans’ algorithm appears to match GEP in 75-80% of cases.
Alternative Algorithm(s)
The University of Nebraska group that originally developed the Hans algorithm has developed a new IHC stain algorithm that reportedly classifies cases of DLBCL more accurately compared to the corresponding molecular subtypes (~80% concordance). This algorithm uses GCET1, CD10, BCL-6, MUM-1, and FOXP1 with differing cutoff values for positive/negative. (WW Choi, et al) New algorithms with IHC markers not commonly used in many laboratories has probably limited popularity compared to the Hans’ algorithm. The 2016 WHO hematopathology revision requires that cases of DLBCL be characterized at GCB vs. non-GCB by some acceptable methodology (molecular or IHC).
References
Hans CP, Weisenburger DD, Greiner TC, Gascoyne RD, Delabie J, Ott G, et al. Confirmation of the molecular classification of diffuse large B-cell lymphoma by immunohistochemistry using a tissue microarray. Blood. 2004;103: 275–282. doi:10.1182/blood-2003-05-1545
Haarer CF, Roberts RA, Frutiger YM, Grogan TM, Rimsza LM. Immunohistochemical classification of de novo, transformed, and relapsed diffuse large B-cell lymphoma into germinal center B-cell and nongerminal center B-cell subtypes correlates with gene expression profile and patient survival. Arch Pathol Lab Med. 2006;130: 1819–1824.
Chang C-C, McClintock S, Cleveland RP, Trzpuc T, Vesole DH, Logan B, et al. Immunohistochemical expression patterns of germinal center and activation B-cell markers correlate with prognosis in diffuse large B-cell lymphoma. Am J Surg Pathol. 2004;28: 464–470.
Choi WWL, Weisenburger DD, Greiner TC, Piris MA, Banham AH, Delabie J, et al. A new immunostain algorithm classifies diffuse large B-cell lymphoma into molecular subtypes with high accuracy. Clin Cancer Res. 2009;15: 5494–5502. doi:10.1158/1078-0432.CCR-09-0113
Alizadeh AA, Elsen MB, Davis RE, Ma C. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 2000.
Swerdlow SH, Campo E, Pileri SA, Harris NL, Stein H, Siebert R, et al. The 2016 revision of the World Health Organization classification of lymphoid neoplasms. Blood. 2016;127: 2375–2390. doi:10.1182/blood-2016-01-643569